Note
Go to the end to download the full example code.
Linear and non-linear ESMDA examples#
A basic example of ESMDA using a simple 1D equation.
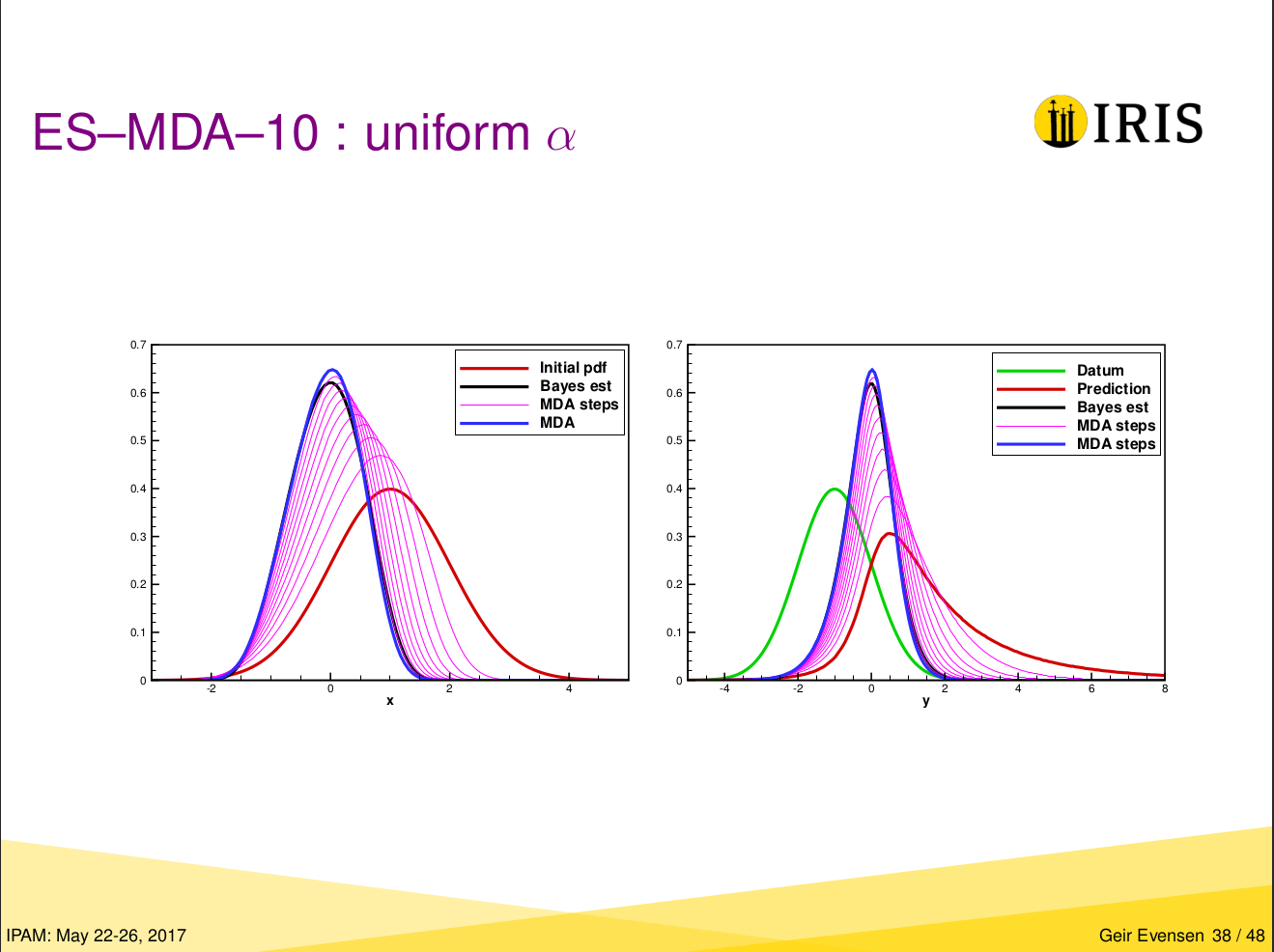
Geir Evensen gave a talk on Properties of Iterative Ensemble Smoothers and Strategies for Conditioning on Production Data at the IPAM in May 2017.
Here we reproduce the examples he showed on pages 34 and 38. The material can be found at:
PDF: http://helper.ipam.ucla.edu/publications/oilws3/oilws3_14079.pdf
Video can be found here: https://www.ipam.ucla.edu/programs/workshops/workshop-iii-data-assimilation-uncertainty-reduction-and-optimization-for-subsurface-flow/?tab=schedule
Geir gives the ESMDA equations as
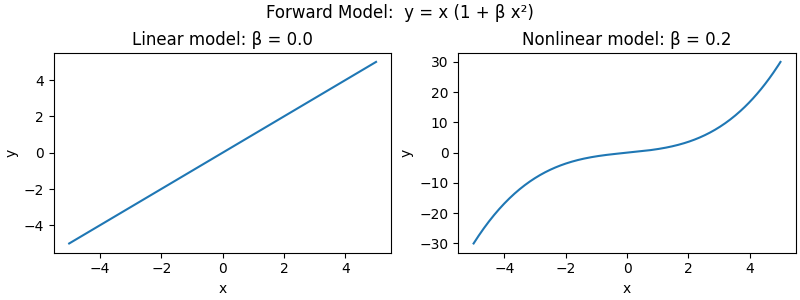
The model used for this example is
which is a linear model if \(\beta=0\).
import numpy as np
import matplotlib.pyplot as plt
import dageo
Forward model#
def forward(x, beta):
"""Simple model: y = x (1 + β x²) (linear if beta=0)."""
return np.atleast_1d(x * (1 + beta * x**2))
fig, axs = plt.subplots(
1, 2, figsize=(8, 3), sharex=True, constrained_layout=True)
fig.suptitle("Forward Model: y = x (1 + β x²)")
px = np.linspace(-5, 5, 301)
for i, b in enumerate([0.0, 0.2]):
axs[i].set_title(
f"{['Linear model', 'Nonlinear model'][i]}: β = {b}")
axs[i].plot(px, forward(px, b))
axs[i].set_xlabel('x')
axs[i].set_ylabel('y')
Plotting functions#
def pseudopdf(data, bins=200, density=True, **kwargs):
"""Return the pdf from a simple bin count.
If the data contains a lot of samples, this should be "smooth" enough - and
much faster than estimating the pdf using, e.g.,
`scipy.stats.gaussian_kde`.
"""
x, y = np.histogram(data, bins=bins, density=density, **kwargs)
return (y[:-1]+y[1:])/2, x
def plot_result(mpost, dpost, dobs, title, ylim):
"""Wrapper to use the same plotting for the linear and non-linear case."""
fig, (ax1, ax2) = plt.subplots(
1, 2, figsize=(10, 4), sharey=True, constrained_layout=True)
fig.suptitle(title)
# Plot Likelihood
ax2.plot(
*pseudopdf(dageo.rng.normal(dobs, size=(ne, dobs.size))),
'C2', lw=2, label='Datum'
)
# Plot steps
na = mpost.shape[0]-1
for i in range(na+1):
params = {
'color': 'C0' if i == na else 'C3', # Last blue, rest red
'lw': 2 if i in [0, na] else 1, # First/last thick
'alpha': 1 if i in [0, na] else i/na, # start faint
'label': ['Initial', *((na-2)*('',)), 'MDA steps', 'MDA'][i],
}
ax1.plot(*pseudopdf(mpost[i, :, 0], range=(-3, 5)), **params)
ax2.plot(*pseudopdf(dpost[i, :, 0], range=(-5, 8)), **params)
# Axis and labels
ax1.set_title('Model Parameter Domain')
ax1.set_xlabel('x')
ax1.set_ylim(ylim)
ax1.set_xlim([-3, 5])
ax1.legend()
ax2.set_title('Data Domain')
ax2.set_xlabel('y')
ax2.set_xlim([-5, 8])
ax2.legend()
Linear case#
Prior model parameters and ESMDA parameters#
In reality, the prior would be \(j\) models provided by, e.g., the geologists. Here we create $j$ realizations using a normal distribution of a defined mean and standard deviation.
# Point of our "observation"
xlocation = -1.0
# Ensemble size
ne = int(1e7)
# Data standard deviation: ones (for this scenario)
obs_std = 1.0
# Prior: Let's start with ones as our prior guess
mprior = dageo.rng.normal(loc=1.0, scale=obs_std, size=(ne, 1))
Run ESMDA and plot#
ESMDA step 1; α=10.0
ESMDA step 2; α=10.0
ESMDA step 3; α=10.0
ESMDA step 4; α=10.0
ESMDA step 5; α=10.0
ESMDA step 6; α=10.0
ESMDA step 7; α=10.0
ESMDA step 8; α=10.0
ESMDA step 9; α=10.0
ESMDA step 10; α=10.0
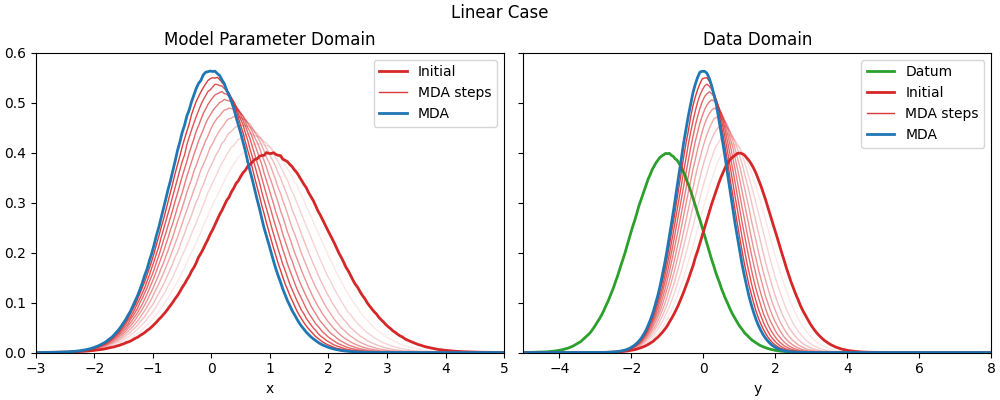
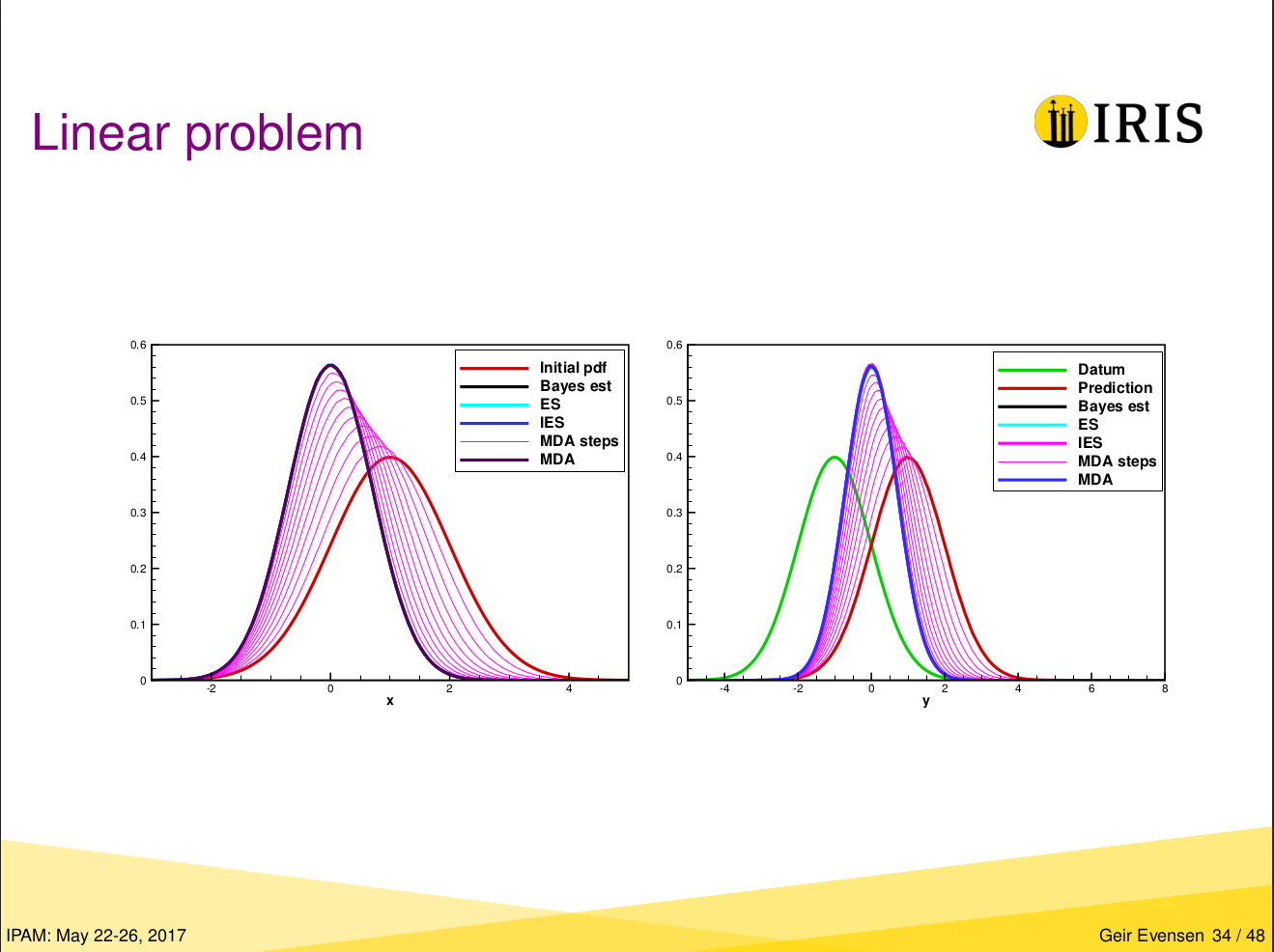
Original figure from Geir’s presentation#
Nonlinear case#
def nonlin_fwd(x):
"""Nonlinear forward model."""
return forward(x, beta=0.2)
# Sample a nonlinear observation; the rest of the parameters remains the same.
n_dobs = nonlin_fwd(xlocation)
nm_post, nd_post = dageo.esmda(
model_prior=mprior,
forward=nonlin_fwd,
data_obs=n_dobs,
sigma=obs_std,
alphas=10,
return_steps=True,
)
ESMDA step 1; α=10.0
ESMDA step 2; α=10.0
ESMDA step 3; α=10.0
ESMDA step 4; α=10.0
ESMDA step 5; α=10.0
ESMDA step 6; α=10.0
ESMDA step 7; α=10.0
ESMDA step 8; α=10.0
ESMDA step 9; α=10.0
ESMDA step 10; α=10.0
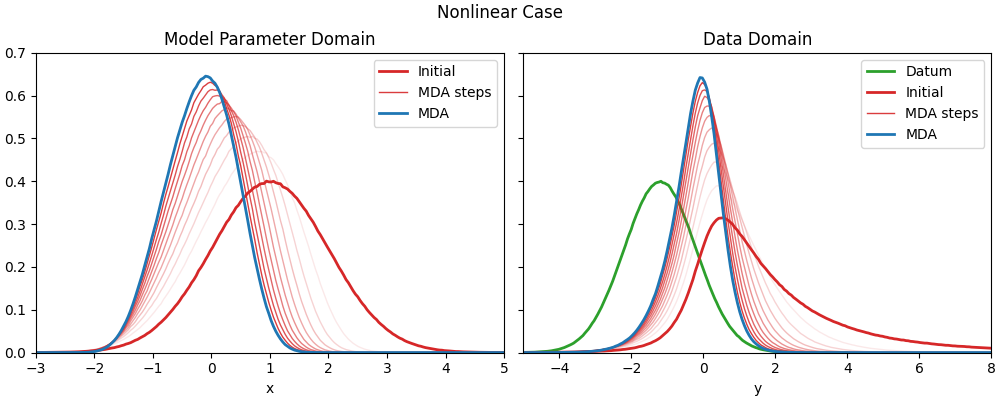
Original figure from Geir’s presentation#
dageo.Report()
Total running time of the script: (0 minutes 12.180 seconds)
Estimated memory usage: 4038 MB