Note
Go to the end to download the full example code.
Geothermal Case Study#
ESMDA example predicting temperature at a production well as a function of permeability.
This example demonstrates the application of the Ensemble Smoother with
Multiple Data Assimilation (ESMDA) using the dageo library to predict
temperature at a production well in a geothermal reservoir. The notebook
integrates the Delft Advanced Research Terra Simulator (DARTS) to model the impact of permeability
variations on temperature over a 30-year period. The example uses a channelized
permeability field, which provides an interesting case study of ESMDA’s
behavior with non-Gaussian geological features. Since ESMDA operates under
Gaussian assumptions, it tends to create smooth updates to the permeability
field rather than maintaining sharp channel boundaries. This limitation becomes
particularly visible when the algorithm identifies the need for connectivity
between wells - instead of creating or modifying channels, it increases
permeability in a more diffuse manner. This behavior highlights both the power
of ESMDA in matching production data and its limitations in preserving complex
geological features.
Tip
This example can serve as an example how one can use dageo with any
external modelling code, here with the Delft Advanced Research Terra
Simulator (DARTS).
Thanks to Mona Devos, who provided the initial version of this example!
Pre-computed data#
DARTS computation#
The DARTS computation in this example takes too long for it to be run
automatically on GitHub with every commit for the documentation (CI). We
therefore pre-computed the results locally to show them here. You can change
the flag compute_darts from False to True to actually compute the
results with DARTS yourself. The pre-computed results were generated with the
following versions (taking roughly 4 h on a single thread):
open-darts: 1.1.4
Python: 3.10.15
NumPy: 2.0.2
SciPy: 1.14.1
Fluvial models#
The fluvial models were generated with FLUVSIM through geomodpy, for
more information see towards the end of the example where the code is shown to
reproduce the facies.
Important
To retrieve the pre-computed data (for both DARTS and the facies), you need
to have pooch installed:
pip install pooch
or
conda install -c conda-forge pooch
import pooch
import numpy as np
import matplotlib.pyplot as plt
import dageo
compute_darts = False
if compute_darts:
from darts.engines import redirect_darts_output
from darts.models.darts_model import DartsModel
from darts.physics.geothermal.physics import Geothermal
from darts.reservoirs.struct_reservoir import StructReservoir
from darts.physics.geothermal.property_container import PropertyContainer
redirect_darts_output("run_geothermal.log")
# For reproducibility, we instantiate a random number generator with a
# fixed seed. For production, remove the seed!
rng = np.random.default_rng(42)
Load pre-computed data#
folder = "data"
ffacies = "facies_geothermal.npy"
fdarts = "darts_output_geothermal.npz"
# Load Facies: Not needed if you compute the facies yourself,
# as described at the end of the notebook.
fpfacies = pooch.retrieve(
"https://raw.github.com/tuda-geo/data/2024-11-30/resmda/"+ffacies,
"9b18f1c80aea93d7973effafde001aa7e72a21ac91edf08e3899d5486998ad2e",
fname=ffacies,
path=folder,
)
facies = np.load(fpfacies)
# Load pre-computed DARTS result; only needed if `compute_darts=False`.
if not compute_darts:
fpdarts = pooch.retrieve(
"https://raw.github.com/tuda-geo/data/2024-11-30/resmda/"+fdarts,
"5622729cd5dc7214de8a199512ace39bda48bff113b2eddb0c48593a57c020d1",
fname=fdarts,
path=folder,
)
pc_darts = np.load(fpdarts)
Convert facies to permeability#
Here we define some model parameters. Important: These have obviously to agree with the facies you computed!
# Model parameters
nx, ny, nz = 60, 60, 3 # 60 x 60 x 3 cells
dx, dy, dz = 30, 30, 30 # Each cell is a voxel of 30 x 30 x 30 meter
ne = 100 # 100 ensembles
years = np.arange(31) # Time: we are modelling 30 years:
# Well locations
iw = [30, 30]
jw = [14, 46]
# Minimum and maximum values for permeability
perm_min = 100.
perm_max = 200.
# Get permeability fields by populating the facies
perm = np.zeros(facies.shape)
perm[facies == 0] = perm_min # outside channels minimum
perm[facies > 0] = perm_max # inside channels maximum
# We use a model with 3 layers, starting all with the same permeability.
perm = np.stack([perm]*nz, axis=-1) # 3x the same
# Assign true permeability (first) and prior permeability
perm_true = perm[:1, :, :, :]
perm_prior = perm[1:, :, :, :]
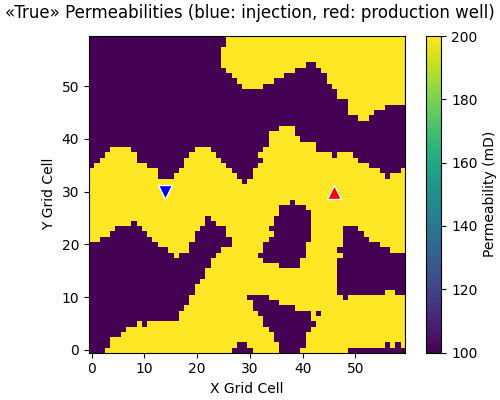
Plot “true” model#
The true permeability model is the one facies realization we use to create observations by adding noise to the modelled data. These observations are what we try to match with ESMDA. To look at this permeability field is important to later interpret the result. In particular, we see that there is a channel of high permeability connecting the injection with the production well.
fopts = {"sharex": True, "sharey": True, "constrained_layout": True}
popts = {"vmin": perm_min, "vmax": perm_max, "origin": "lower"}
fig, ax = plt.subplots(figsize=(5, 4), **fopts)
# Permeabilities
im = ax.imshow(perm_true[0, :, :, 0], **popts)
# Wells
ax.plot(jw[0], iw[0], "v", c="b", ms=10, mec="w")
ax.plot(jw[1], iw[1], "^", c="r", ms=10, mec="w")
# Labels and colour bar
ax.set_ylabel("Y Grid Cell")
ax.set_xlabel("X Grid Cell")
fig.suptitle(
"«True» Permeabilities (blue: injection, red: production well)"
)
cbar = fig.colorbar(im, ax=ax, label="Permeability (mD)")
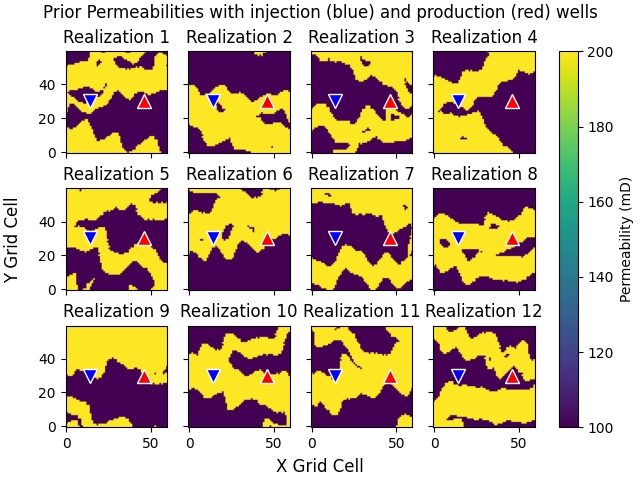
Prior permeabilities#
Plot the first 12 prior permeability models; yellow shows the channels with higher permeability.
fig, axs = plt.subplots(3, 4, **fopts)
for i, ax in enumerate(axs.ravel()):
ax.set_title(f"Realization {i+1}")
# Permeabilities
im = ax.imshow(perm_prior[i, :, :, 0], **popts)
# Wells
ax.plot(jw[0], iw[0], "v", c="b", ms=10, mec="w")
ax.plot(jw[1], iw[1], "^", c="r", ms=10, mec="w")
# Labels and colour bar
fig.supylabel("Y Grid Cell")
fig.supxlabel("X Grid Cell")
fig.suptitle(
"Prior Permeabilities with injection (blue) and production (red) wells"
)
cbar = fig.colorbar(im, ax=axs.ravel(), label="Permeability (mD)")
Simulate prior data#
Here we simulate “true” and prior data, and create “observed” data from adding random noise to the true data.
if compute_darts:
data_true = temperature_at_production_well(perm_true)
data_prior = temperature_at_production_well(perm_prior)
# Add noise to the true data and create observed data
dstd = 0.2
data_obs = rng.normal(data_true, dstd)
else:
data_true = pc_darts["data_true"].astype("d")
data_prior = pc_darts["data_prior"].astype("d")
data_obs = pc_darts["data_obs"].astype("d")
k2c = -273.15 # To plot °C instead of K
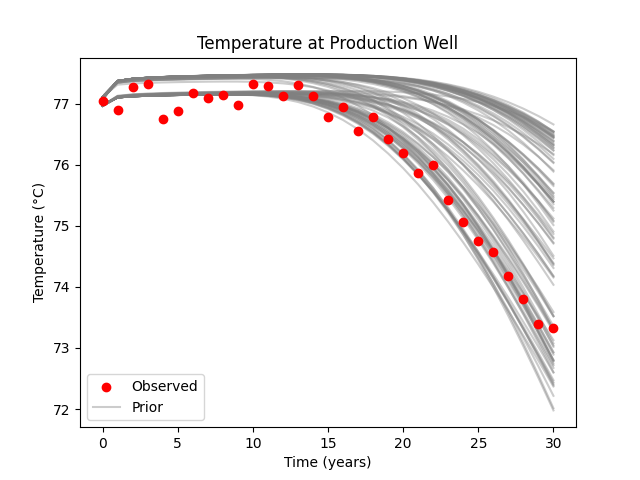
Plot prior data#
The prior data show that there seem to be two possible clusters, one having slightly higher temperatures than the other. The slightly colder cluster corresponds to the models that have a connecting channel, the slightly warmer cluster the models that have no connecting channel.
fig, ax = plt.subplots()
ax.scatter(years, data_obs+k2c, label="Observed", c="r", zorder=10)
ax.plot(years, data_prior.T+k2c, c="grey", alpha=0.4,
label=["Prior"] + [None] * (ne - 1))
ax.legend()
ax.set_xlabel("Time (years)")
ax.set_ylabel("Temperature (°C)")
ax.set_title("Temperature at Production Well")
Perform Data Assimilation (ESMDA)#
# Define a function to restrict the permeability values
def restrict_permeability(x):
"""Restrict possible permeabilities."""
np.clip(x, perm_min, perm_max, out=x)
# Run ESMDA
if compute_darts:
perm_post, data_post = dageo.esmda(
model_prior=perm_prior,
forward=temperature_at_production_well,
data_obs=data_obs,
sigma=dstd,
alphas=4,
data_prior=data_prior,
callback_post=restrict_permeability,
random=rng,
)
# Store result
np.savez_compressed(
file="darts_output_geothermal.npz",
data_true=data_true.astype("f"),
data_obs=data_obs.astype("f"),
data_prior=data_prior.astype("f"),
data_post=data_post.astype("f"),
perm_post=perm_post.astype("f"),
allow_pickle=False,
)
else:
data_post = pc_darts["data_post"].astype("d")
perm_post = pc_darts["perm_post"].astype("d")
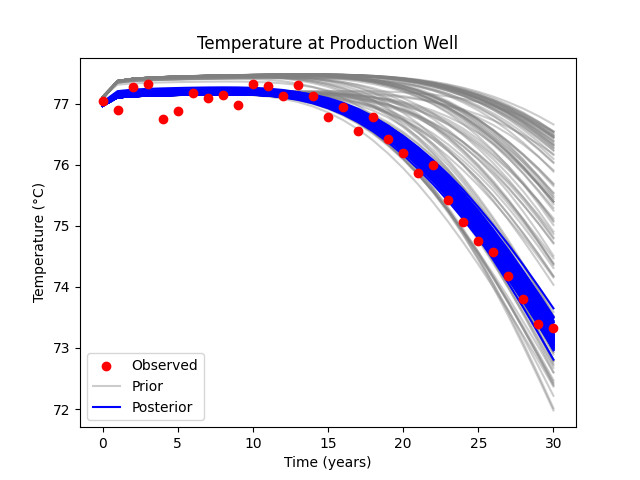
Plot posterior data#
Models that connect the injection and production well with higher permeability (lower temperature) are the ones which match the data better. Even when we originally did not have channels between the injection and production well, the model found that the best way to match the data is to increase the permeability between the wells.
fig, ax = plt.subplots()
ax.scatter(years, data_obs+k2c, label="Observed", c="r", zorder=10)
ax.plot(years, data_prior.T+k2c, c="grey", alpha=0.4,
label=["Prior"] + [None] * (ne - 1))
ax.plot(years, data_post.T+k2c, c="b",
label=["Posterior"] + [None] * (ne - 1))
ax.legend()
ax.set_xlabel("Time (years)")
ax.set_ylabel("Temperature (°C)")
ax.set_title("Temperature at Production Well")
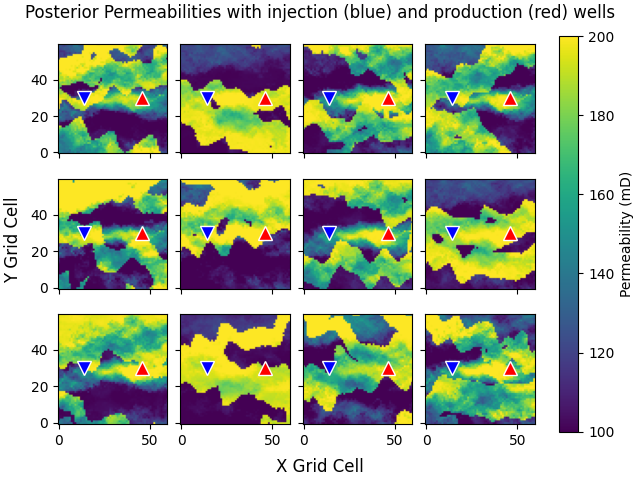
Plot posterior permeabilities#
Plot the first 12 posterior permeability models.
The data is matched nicely (above figure), yet we still have different realizations, meaning that the posterior does not look like the “true” model, as expected (ill-posed problem). This means that we still have uncertainty in the model, and also that the ensemble has not collapsed to a single model.
fig, axs = plt.subplots(3, 4, **fopts)
axs = axs.ravel()
for i in range(min(ne, 12)):
im = axs[i].imshow(perm_post[i, :, :, 0], **popts)
axs[i].plot(jw[0], iw[0], "v", c="b", ms=10, mec="w")
axs[i].plot(jw[1], iw[1], "^", c="r", ms=10, mec="w")
fig.colorbar(im, ax=axs, label="Permeability (mD)")
fig.suptitle(
"Posterior Permeabilities with injection (blue) and production (red) wells"
)
fig.supylabel("Y Grid Cell")
fig.supxlabel("X Grid Cell")
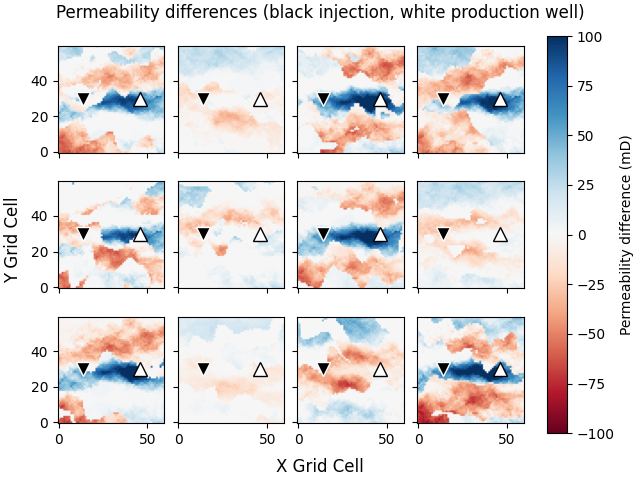
Plot permeability differences#
The difference between prior and posterior shows nicely that in models, where there is a channel between the injection and production well, not much had to change in terms of permeability to predict the data. In models, however, where there is no connecting channel, it increased significantly the permeability between the wells, and decreased them further out.
popts = {"vmin": -100, "vmax": 100, "origin": "lower", "cmap": "RdBu"}
fig, axs = plt.subplots(3, 4, **fopts)
axs = axs.ravel()
for i in range(min(ne, 12)):
im = axs[i].imshow(perm_post[i, :, :, 0] - perm_prior[i, :, :, 0], **popts)
axs[i].plot(jw[0], iw[0], "v", c="k", ms=10, mec="w")
axs[i].plot(jw[1], iw[1], "^", c="w", ms=10, mec="k")
fig.colorbar(im, ax=axs, label="Permeability difference (mD)")
fig.suptitle(
"Permeability differences (black injection, white production well)"
)
fig.supylabel("Y Grid Cell")
fig.supxlabel("X Grid Cell")
Reproduce the facies#
Note
The following cell is about how to reproduce the facies data loaded in
facies_geothermal.nc. This was created using geomodpy.
geomodpy (Guillaume Rongier, 2023) is not open-source yet. The
functionality of geomodpy that we use here is a python wrapper for the
Fortran library FLUVSIM published in:
Deutsch, C. V., and T. T. Tran, 2002, FLUVSIM: a program for object-based stochastic modeling of fluvial depositional systems: Computers & Geosciences, 28(4), 525–535.
# FLUVSIM Version used: 2.900
from geomodpy.wrapper.fluvsim import FLUVSIM
# Dimensions
nx, ny = 60, 60
dx, dy = 30, 30
ne = 100
# Create a fluvsim instance with the geological parameters
fluvsim = FLUVSIM(
channel_orientation=(60., 90., 120.),
# Proportions for each facies
channel_prop=0.5,
crevasse_prop=0.0,
levee_prop=0.0,
# Parameters defining the grid
shape=(nx, ny),
spacing=(dx, dy),
origin=(0, 0),
# Number of realizations and random seed
n_realizations=ne+1, # +1 for "true"
seed=42,
)
# Run fluvsim to create the facies
facies = fluvsim.run()
facies = facies.data_vars["facies code"].values.astype("i4")
# Save the facies values as a compressed npy-file.
np.save("facies_geothermal.npy", facies.squeeze(), allow_pickle=False)
dageo.Report()
Total running time of the script: (0 minutes 5.232 seconds)
Estimated memory usage: 133 MB