Note
Go to the end to download the full example code.
Localization#
Example using several well doublets and compares ESMDA with and without localization.
The example follows contextually 2D Reservoir ESMDA example.
import numpy as np
import matplotlib.pyplot as plt
import dageo
# For reproducibility, we instantiate a random number generator with a fixed
# seed. For production, remove the seed!
rng = np.random.default_rng(2020)
Model parameters#
# Grid extension
nx = 35
ny = 30
nc = nx*ny
# Permeabilities
perm_mean = 3.0
perm_min = 0.5
perm_max = 5.0
# ESMDA parameters
ne = 100 # Number of ensembles
dt = np.zeros(10)+0.0001 # Time steps (could be irregular, e.g., increasing!)
time = np.r_[0, np.cumsum(dt)]
# Assumed sandard deviation of our data
dstd = 0.5
# Measurement points
ox = (5, 15, 24)
oy = (5, 10, 24)
# Number of data points
nd = time.size * len(ox)
# Wells
wells = np.array([
[ox[0], oy[0], 180], [5, 12, 120],
[ox[1], oy[1], 180], [20, 5, 120],
[ox[2], oy[2], 180], [24, 17, 120]
])
Create permeability maps for ESMDA#
We will create a set of permeability maps that will serve as our initial guess (prior). These maps are generated using a Gaussian random field and are constrained by certain statistical properties.
# Get the model and ne prior models
RP = dageo.RandomPermeability(nx, ny, perm_mean, perm_min, perm_max)
perm_true = RP(1, random=rng)
perm_prior = RP(ne, random=rng)
Run the prior models and the reference case#
# Instantiate reservoir simulator
RS = dageo.Simulator(nx, ny, wells=wells)
def sim(x):
"""Custom fct to use exp(x), and specific dt & location."""
return RS(np.exp(x), dt=dt, data=(ox, oy)).reshape((x.shape[0], -1))
# Simulate data for the prior and true fields
data_prior = sim(perm_prior)
data_true = sim(perm_true)
data_obs = rng.normal(data_true, dstd)
data_obs[0, :3] = data_true[0, :3]
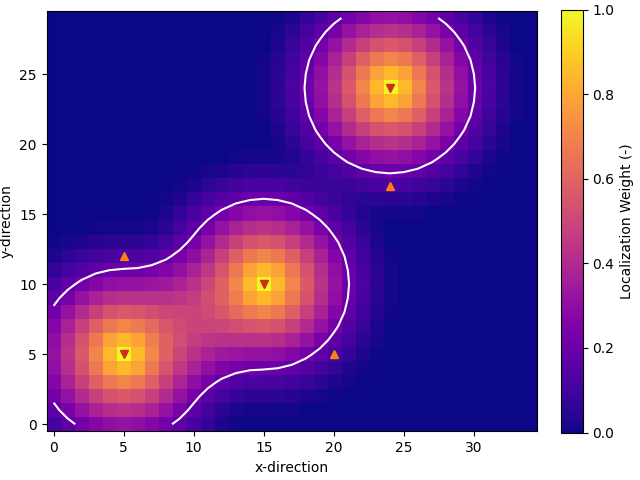
Localization Matrix#
# Vector of nd length with the well x and y position for each nd data point
nd_positions = np.tile(np.array([ox, oy]), time.size).T
# Create matrix
loc_mat = dageo.localization_matrix(RP.cov, nd_positions, (nx, ny))
# QC localization matrix
fig, ax = plt.subplots(1, 1, constrained_layout=True)
im = ax.imshow(loc_mat.sum(axis=2).T/time.size, origin='lower', cmap="plasma")
ax.contour(loc_mat.sum(axis=2).T/time.size, levels=[0.2, ], colors='w')
ax.set_xlabel('x-direction')
ax.set_ylabel('y-direction')
for well in wells:
ax.plot(well[0], well[1], ['C3v', 'C1^'][int(well[2] == 120)])
fig.colorbar(im, ax=ax, label="Localization Weight (-)")
ESMDA#
def restrict_permeability(x):
"""Restrict possible permeabilities."""
np.clip(x, perm_min, perm_max, out=x)
inp = {
'model_prior': perm_prior,
'forward': sim,
'data_obs': data_obs,
'sigma': dstd,
'alphas': 4,
'data_prior': data_prior,
'callback_post': restrict_permeability,
'random': rng,
}
Without localization#
nl_perm_post, nl_data_post = dageo.esmda(**inp)
ESMDA step 1; α=4.0
ESMDA step 2; α=4.0
ESMDA step 3; α=4.0
ESMDA step 4; α=4.0
With localization#
wl_perm_post, wl_data_post = dageo.esmda(**inp, localization_matrix=loc_mat)
ESMDA step 1; α=4.0
ESMDA step 2; α=4.0
ESMDA step 3; α=4.0
ESMDA step 4; α=4.0
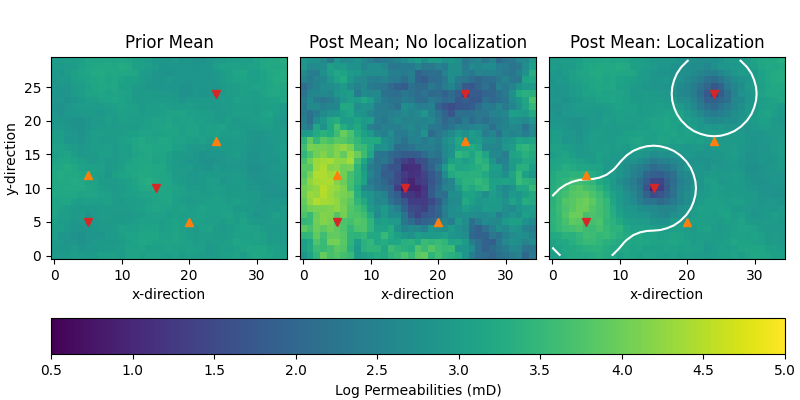
Compare permeabilities#
# Plot posterior
fig, axs = plt.subplots(
1, 3, figsize=(8, 4), sharex=True, sharey=True, constrained_layout=True)
par = {"vmin": perm_min, "vmax": perm_max, "origin": "lower"}
axs[0].set_title("Prior Mean")
im = axs[0].imshow(perm_prior.mean(axis=0).T, **par)
axs[1].set_title("Post Mean; No localization")
axs[1].imshow(nl_perm_post.mean(axis=0).T, **par)
axs[2].set_title("Post Mean: Localization")
axs[2].imshow(wl_perm_post.mean(axis=0).T, **par)
axs[2].contour(loc_mat.sum(axis=2).T, levels=[2.0, ], colors="w")
fig.colorbar(im, ax=axs, label="Log Permeabilities (mD)",
orientation="horizontal")
for ax in axs:
for well in wells:
ax.plot(well[0], well[1], ["C3v", "C1^"][int(well[2] == 120)])
for ax in axs:
ax.set_xlabel('x-direction')
axs[0].set_ylabel('y-direction')
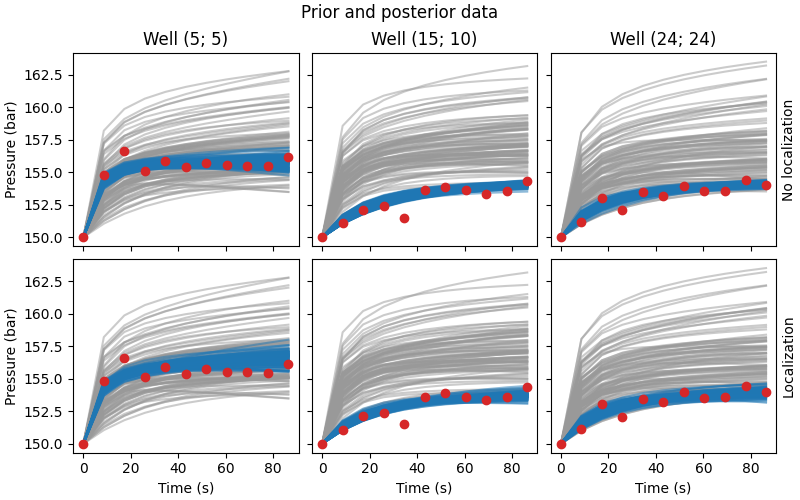
Compare data#
# QC data and priors
fig, axs = plt.subplots(
2, 3, figsize=(8, 5), sharex=True, sharey=True, constrained_layout=True)
fig.suptitle('Prior and posterior data')
for i, ax in enumerate(axs[0, :]):
ax.set_title(f"Well ({ox[i]}; {oy[i]})")
ax.plot(time*24*60*60, data_prior[..., i::3].T, color='.6', alpha=0.5)
ax.plot(time*24*60*60, nl_data_post[..., i::3].T, color='C0', alpha=0.5)
ax.plot(time*24*60*60, data_obs[0, i::3], 'C3o')
for i, ax in enumerate(axs[1, :]):
ax.plot(time*24*60*60, data_prior[..., i::3].T, color='.6', alpha=0.5)
ax.plot(time*24*60*60, wl_data_post[..., i::3].T, color='C0', alpha=0.5)
ax.plot(time*24*60*60, data_obs[0, i::3], 'C3o')
ax.set_xlabel("Time (s)")
for i, ax in enumerate(axs[:, 0]):
ax.set_ylabel("Pressure (bar)")
for i, txt in enumerate(["No l", "L"]):
axs[i, 2].yaxis.set_label_position("right")
axs[i, 2].set_ylabel(f"{txt}ocalization")
dageo.Report()
Total running time of the script: (0 minutes 20.604 seconds)
Estimated memory usage: 131 MB